Multivariate Analysis of Transcript Splicing (MATS)
Xing Lab, University of California, Los Angeles
Updates
|
|
|
|
|
|
|
|
|
Contact
Correspondences regarding the MATS algorithm should be directed to Prof. Yi Xing (yxing at ucla.edu) and Shihao Shen (shihao-shen at uiowa.edu).
Correspondences regarding running of the MATS pipeline should be directed to Juw Won Park (jwpark2012 at ucla.edu).
Citation
Shen S., Park JW., Huang J., Dittmar KA., Lu ZX., Zhou Q., Carstens RP., Xing Y.
MATS: A Bayesian Framework for Flexible Detection of Differential Alternative Splicing from RNA-Seq Data.
Nucleic Acids Research, 2012;40(8):e61 doi: 10.1093/nar/gkr1291
About MATS
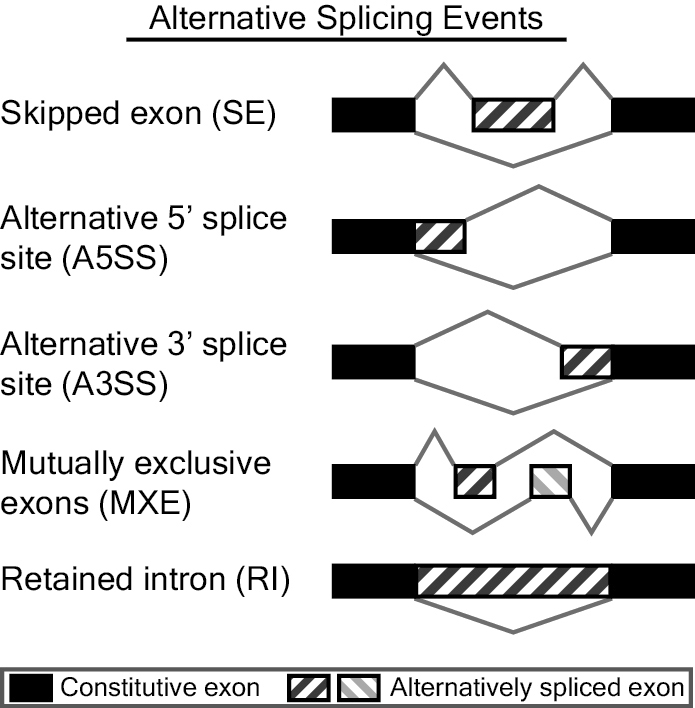
MATS is a computational tool to detect differential alternative splicing events from RNA-Seq data. The statistical model of MATS calculates the P-value and false discovery rate that the difference in the isoform ratio of a gene between two conditions exceeds a given user-defined threshold. From the RNA-Seq data, MATS can automatically detect and analyze alternative splicing events corresponding to all major types of alternative splicing patterns. MATS handles replicate RNA-Seq data from both paired and unpaired study design.
Software Download
|
|
Documentation
Pre-requisites
|
|
|